Home - Research - Software - Workshops - Contact
Research
I develop machine learning methods to solve the many challenges that come along with biomedical image analysis. The sections below provide an overview of the fields where I have contributed.
Image segmentation
Image segmentation is the fundamental task of assigning class labels to each pixel in the image.
- Remmerie, A., Martens, L., Thoné, T., Castoldi, A., Seurnick, R., Pavie, B., Browaeys, R., Roels, J., Vanneste, B., Prijck, S. De, Vanhockerhout, M., Devisscher, L., Hoorens, A., Bonnardel, J., Vandamme, N., Kremer, A., Borghgraef, P., Vlierberghe, H. Van, Lippens, S., … Scott, C. L. (2020). Depletion of resident Kupffer cells in NAFLD leads to increased prevalence of hepatocellular carcinoma. Immunity.
- Roels, J., Hennies, J., Saeys, Y., Philips, W., & Kreshuk, A. (2019). Domain Adaptive Segmentation in Volume Electron Microscopy Imaging. IEEE International Symposium on Biomedical Imaging. https://doi.org/arXiv:1810.09734v1
Domain adaptation
In domain adaptation, we consider a (relatively large) labeled source dataset and a target dataset with no or very few label. The goal is to maximize the predictive performance on the target dataset based on all the available labels.
I developed a new domain adaptive architecture that is able to adapt state-of-the-art encoder-decoder models to alternative target domains. The image below illustrates the effect of no domain adaptation (left), a state-of-the-art two-stream architecture (middle) and our proposed network (right).
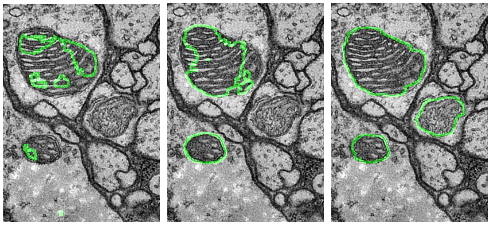
Relevant work:
- Roels, J., Hennies, J., Saeys, Y., Philips, W., & Kreshuk, A. (2019). Domain Adaptive Segmentation in Volume Electron Microscopy Imaging. IEEE International Symposium on Biomedical Imaging. https://doi.org/arXiv:1810.09734v1
Active learning
In active learning situations, we assume the availability of an oracle that can provide labeled data. In each iteration, a set of samples is selected and queried to the oracle, after which the predictor is retrained. The goal is to achieve maximum predictive performance with as few queried samples as possible.
As most active learning methods are designed for image classification, we have adapted the state-of-the-art performing sample selection criteria to image segmentation. The images below show the performance curves of the different proposed active learning methods for segmentation of mitochondria in electron microscopy (EPFL, MiRA, VNC dataset from left to right). We are able to achieve close to fully supervised performance with just 200 to 300 labeled samples.
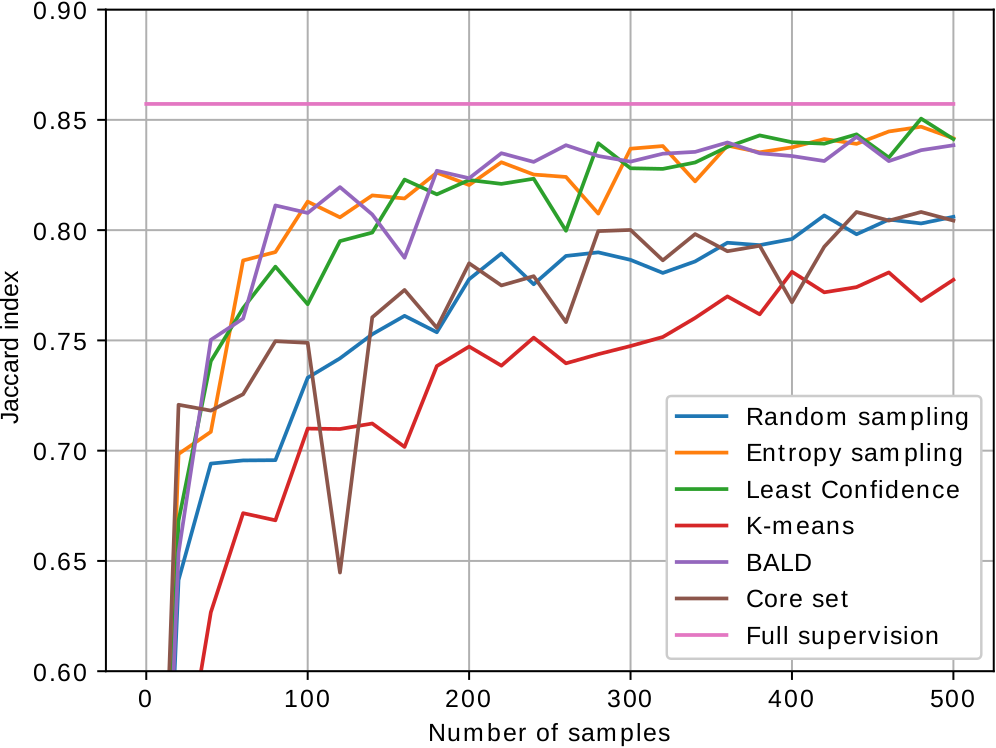
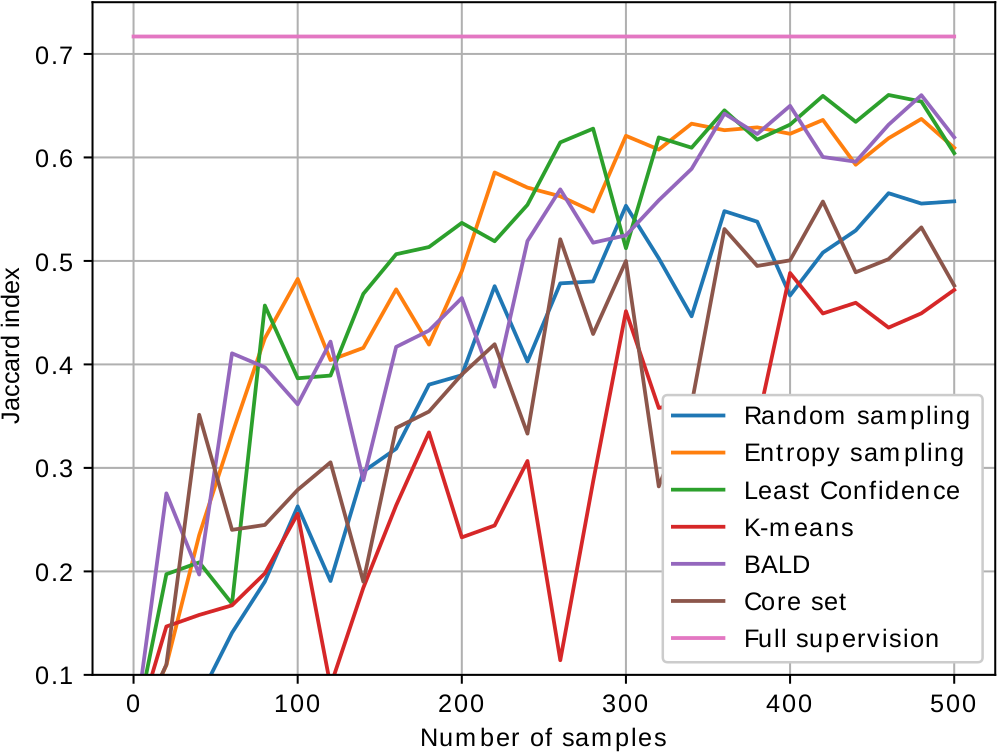
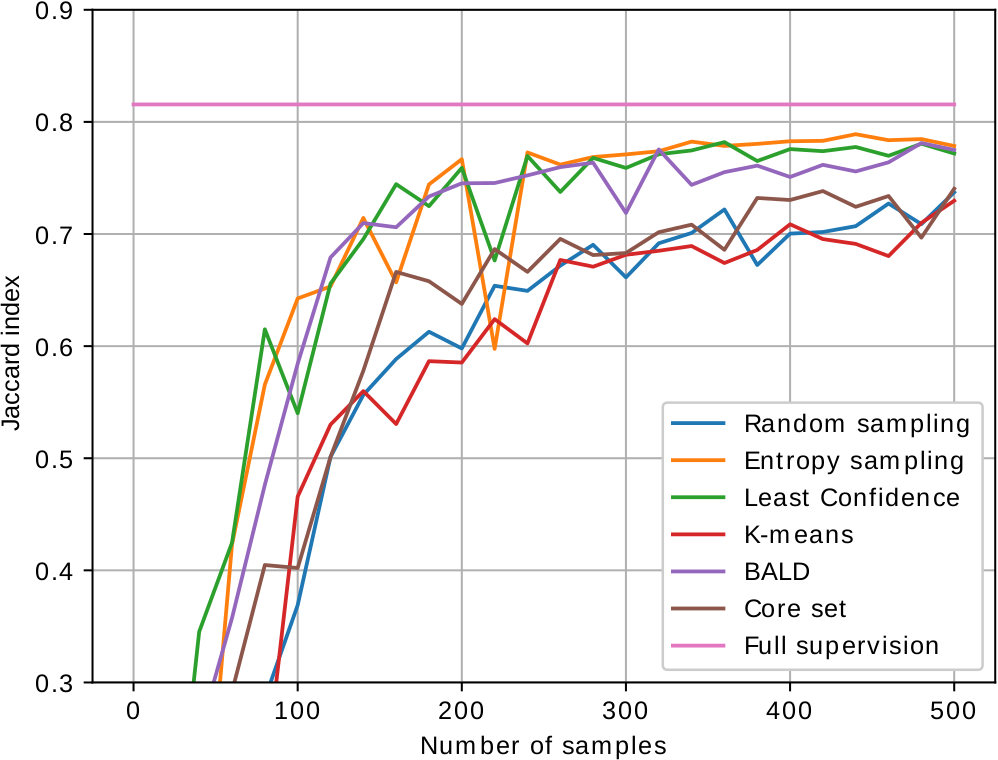
Relevant work:
- Roels, J., & Saeys, Y. (2019). Cost-efficient segmentation of electron microscopy images using active learning. BENELEARN Proceedings.
Image restoration
Image restoration aims at improving the quality of the images by removing noise, blur, etc. This can improve performance of automated analysis techniques in subsequent steps.
I developed state-of-the-art image restoration algorithms that estimate and exploit modality specific parameters in statistical estimation frameworks. The images below - from left to right - show the ground truth high quality image data, the corrupted (noisy and blurred) data, two state-of-the-art image restoration algorithms (respectively tikhonov and non-local deconvolution) and our proposed algorithm. The resulting images from our algorithm are more suitable for analysis and allow faster image acquisition.
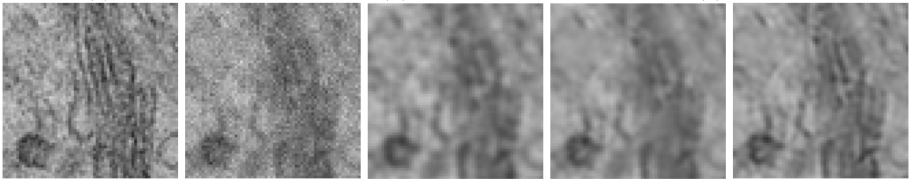
Relevant work:
- Roels, J., Vernaillen, F., Kremer, A., Gonçalves, A., Aelterman, J., Luong, H. Q., Goossens, B., Philips, W., Lippens, S., & Saeys, Y. (2020). An interactive ImageJ plugin for semi-automated image denoising in electron microscopy. Nature Communications, 11(1), 1–13. https://doi.org/10.1038/s41467-020-14529-0
- Roels, J., Aelterman, J., Luong, H. Q., Lippens, S., Pižurica, A., Saeys, Y., Philips, W., Pizurica, A., Saeys, Y., & Philips, W. (2018). An Overview of State-of-the-art Image Restoration in Electron Microscopy. Journal of Microscopy, 271(3), 239–254. https://doi.org/10.1111/jmi.12716
- Roels, J., Aelterman, J., de Vylder, J., Lippens, S., Luong, H. Q., Guérin, C. J., & Philips, W. (2016). Image degradation in microscopic images: Avoidance, artifacts, and solutions. Advances in Anatomy Embryology and Cell Biology, 219(Focus on Bio-image Informatics), 41–67. https://doi.org/10.1007/978-3-319-28549-8_2
- Roels, J., Aelterman, J., De Vylder, J., Luong, H., Saeys, Y., & Philips, W. (2016). Bayesian deconvolution of scanning electron microscopy images using point-spread function estimation and non-local regularization. International Conference of the IEEE Engineering in Medicine and Biology Society, 2016-Octob, 443–447. https://doi.org/10.1109/EMBC.2016.7590735

